Research questions
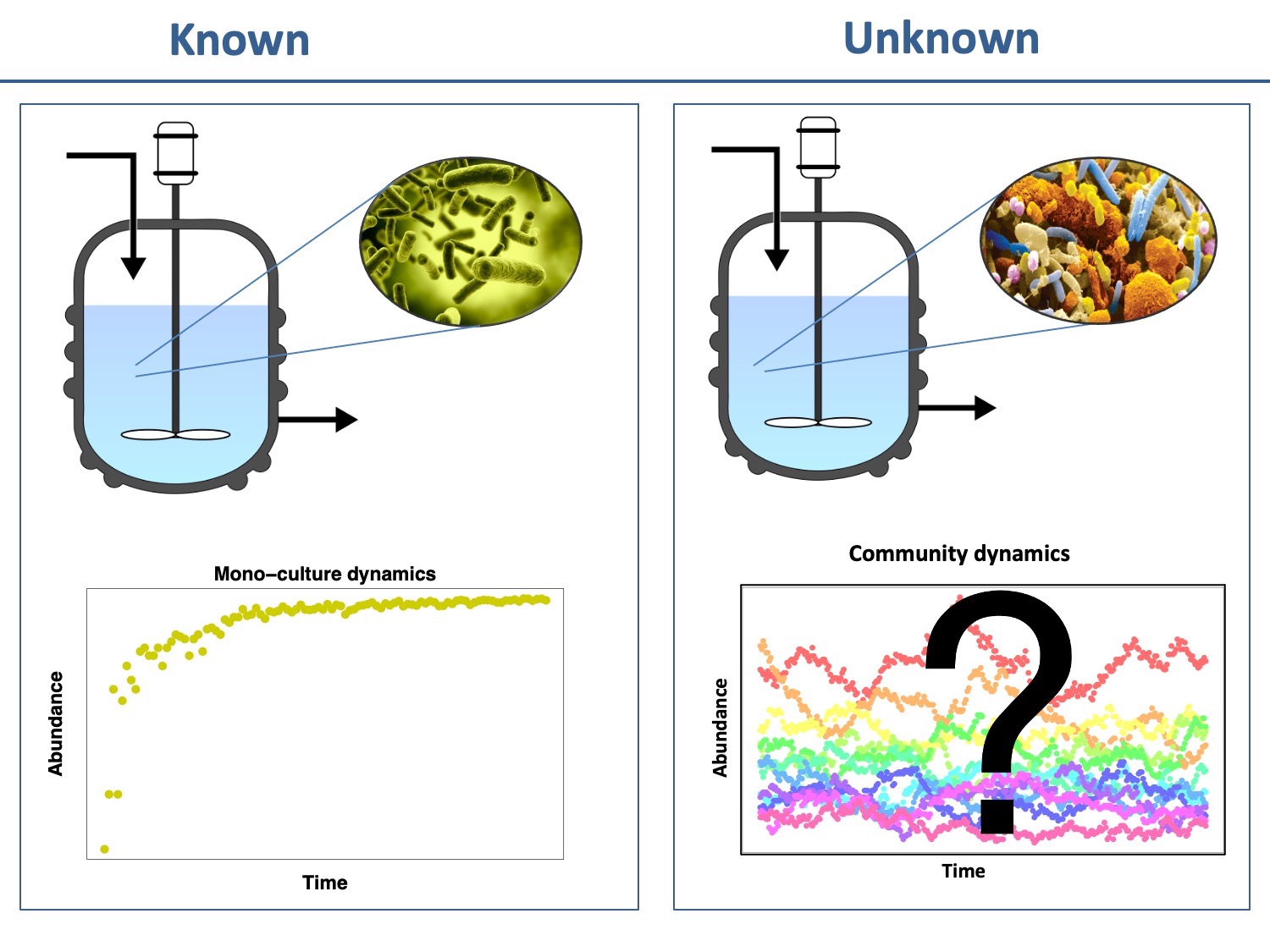
We know quite well what to expect when a single microbial species is grown in a bioreactor. But what happens when we grow several interacting species at once? Will they reach a stable state or fluctuate wildly? Which one(s) will dominate the community? Will it always be the same one(s)? How do perturbations impact the outcome? Do we understand the system well enough that we can predict what will happen? Answering these fundamental questions is important if we want to rationally design microbial communities to accomplish a task, such as improving gut health through probiotic gut bacterial consortia.
Methodology
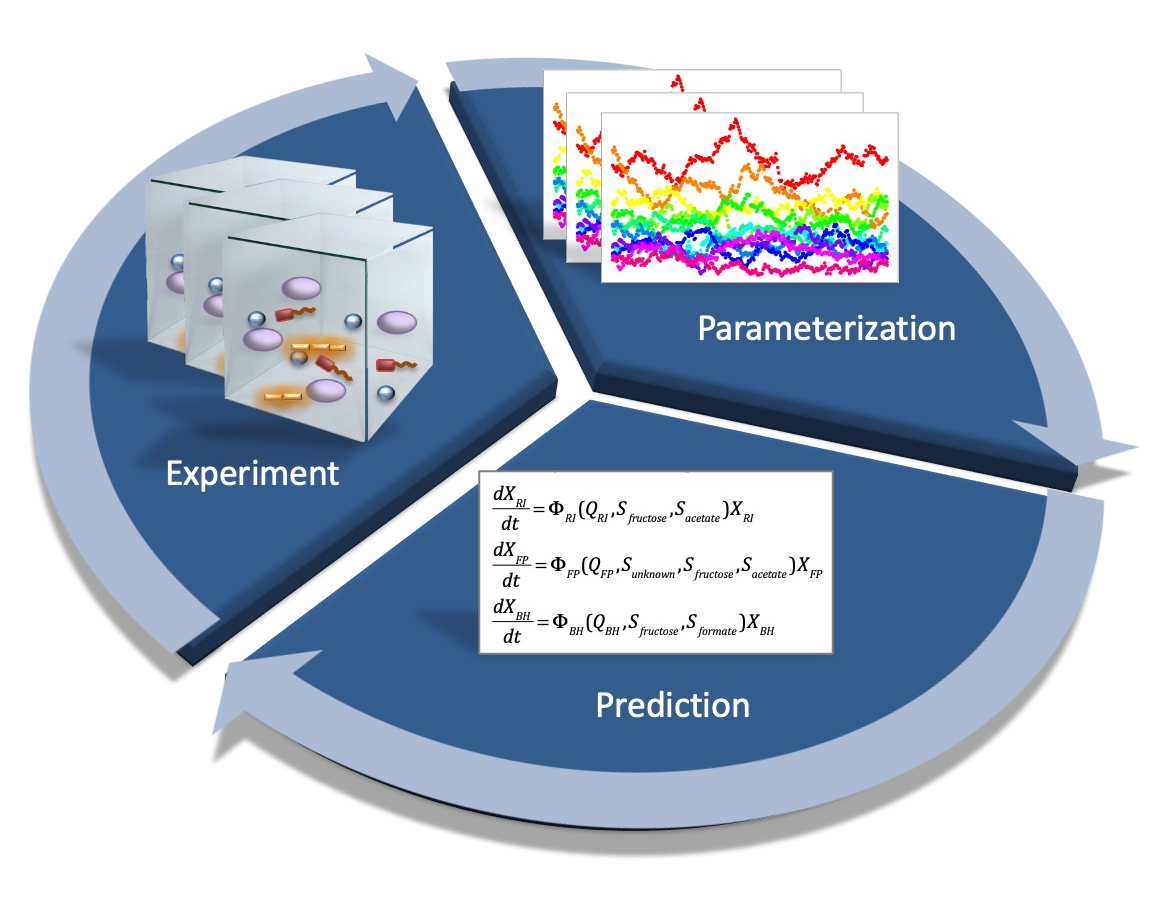
We are investigating these questions with selected bacterial species from the human gut using a systems biology approach. This means that we combine experiments with mathematical modelling. The model describes in abstract terms what we think that the community does. It takes parameters such as consumption and production rates for metabolites, which we measure experimentally. We then test whether the model can reproduce experimental observations or, vice versa, if experiments confirm model predictions. A mismatch tells us about aspects of community dynamics that we do not yet understand and that we need to further explore. In this way, model and experiment inform each other.
Results
A mathematical model for the full community is still under development. To give you an idea which predictions can be made with community models, you can play around with a three-species kinetic model implemented by Daniel Rios Garza.
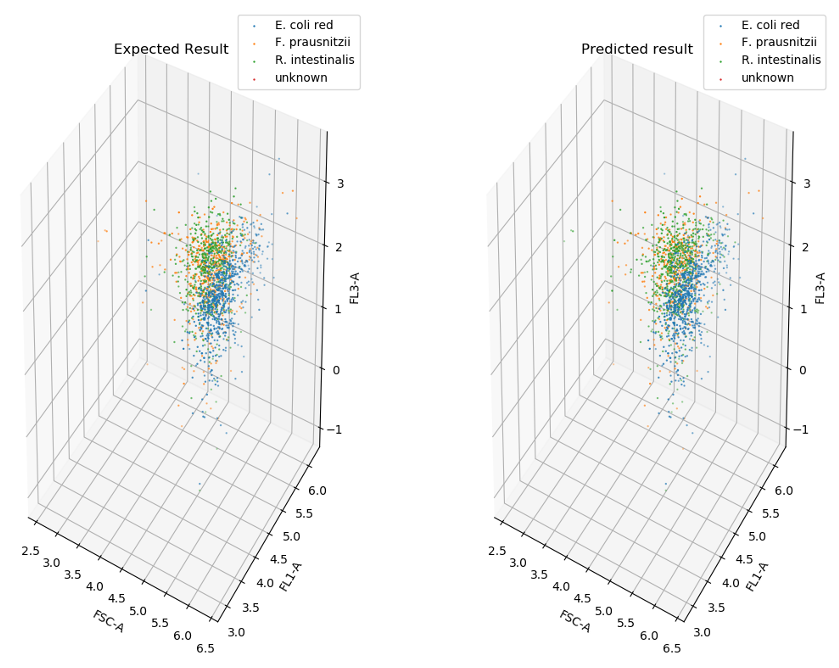
CellScanner
is a tool that uses machine learning to distinguish between cells belonging to different species in flow cytometry data. CellScanner was presented by Clémence Joseph at the ISME summit Unity in Diversity.