CoNet - Documentation - Tutorial (Costello data subset)
The goal of this tutorial is to infer relationships between the oral subset of the Costello data. This tutorial can also be carried out using the "Demo" button in CoNet (but without the metadata).
Tutorial files
oral subset of Costello 2009
metadata accompanying the Costello data
settings file
Short version (using the CoNet settings file)
- Open the CoNet plugin in Cytoscape and click the "Settings loading/saving" button.
- In the Settings menu, click "Select file" and browse the file tree to the settings file.
- Click the "Apply settings in selected file" button and close the Settings menu.
- In the Data menu, click on the top-most "Select file" and browse the file tree to select the Costello data file.
- In the Data menu, click the "Metadata and features" button to open the metadata submenu.
- In the Metadata submenu, click the left "Select file" button and select the metadata file.
- In the main menu, click the GO button.
Long version (entering everything manually in the CoNet interface)
- Open the CoNet plugin in Cytoscape and open the Data menu
- In the Data menu, click on the top-most "Select file" and browse the file tree to select the Costello data file.
- In the Data menu, click the "Metadata and features" button to open the metadata submenu.
- In the Metadata submenu, click the left "Select file" button and select the metadata file.
- In the text area below, enter the metadata columns contained in the metadata file: bodysite/taxon
- Close the Metadata and Data menus and open the Preprocessing menu.
- Select "row_minocc" and set it to 5 in the Input filtering box of the Preprocessing menu.
- Select "col_norm" in the Standardization box of the Preprocessing menu.
- Close the Preprocessing menu and open the Methods menu.
- Select Pearson, Spearman, Bray Curtis and Kullback-Leibler as measures.
- Open the Threshold setting submenu and enter "25" as edge selection parameter.
- Enable "Top and bottom"
- Close the Threshold setting submenu and the Methods menu and open the Merge menu.
- Enable the "multi-graph" option in the Merge menu.
- Close the Merge menu and open the Configuration menu.
- Set the missing value treatment to "pairwise_omit" and the minimum number of NaN-free value pairs to 5.
- In the main menu, click the GO button.
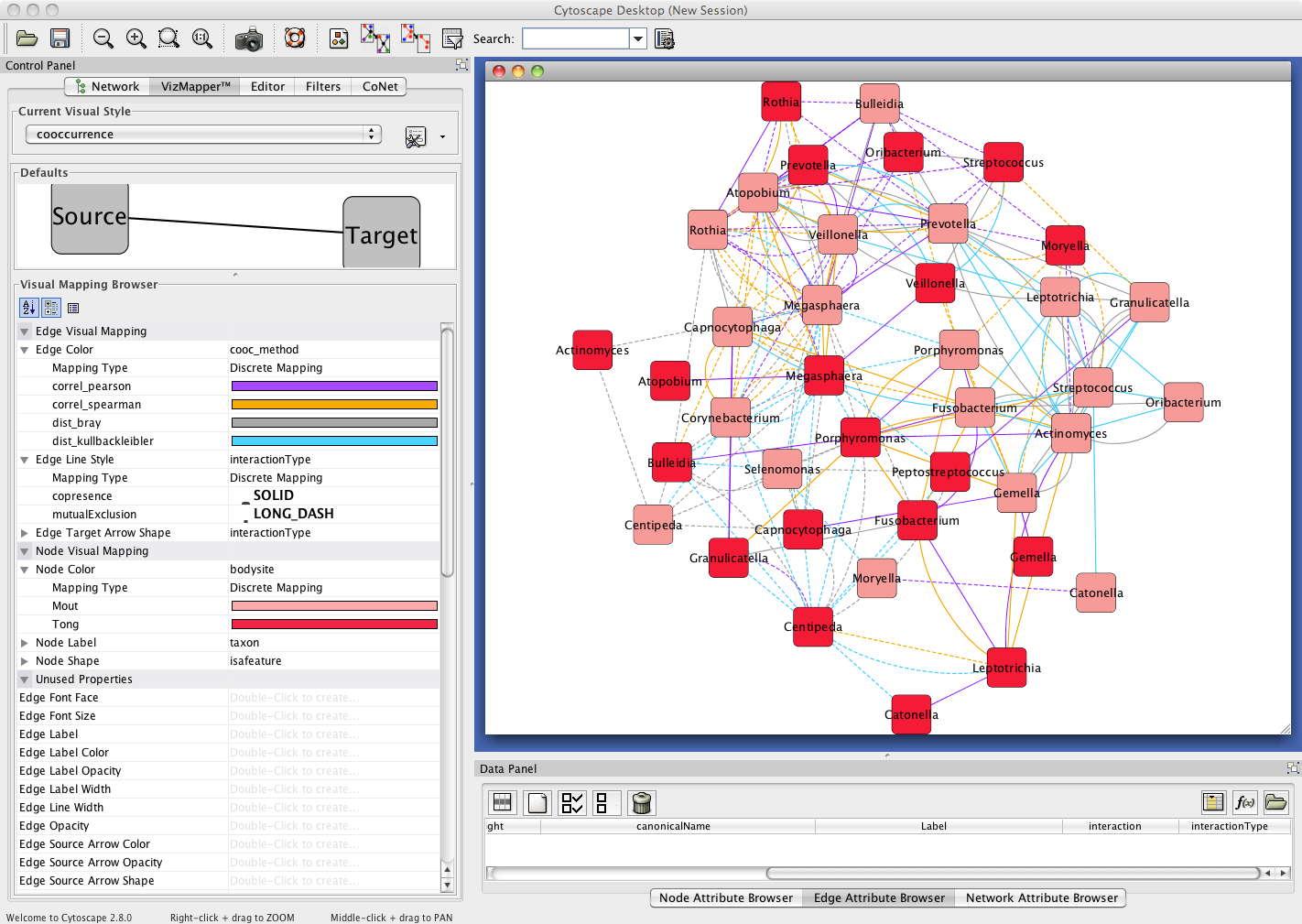
Result
The network returned by CoNet has no layout, as layout computation can be time-consuming, depending on the chosen algorithm.
You can layout the network using one of Cytoscape's supported layouts, for instance the yFiles-layout Organic.
You can then use the VizMapperTM to set the "taxon" node attribute values as node label and to set the node color according
to the "bodysite" node attribute values. The multi-edges can be colored according to their measure using edge attribute "cooc_method" and
their interaction type (attribute "interactionType") can be visualized with solid and dashed lines using the edge line style.
After these processing steps, the network looks like this:
You can save the current CoNet setting by clicking on the "Settings loading/saving" button in the main menu and then selecting a folder and typing a file name to which to save the current settings.