MENA
MENA stands for Molecular Ecological Network Analysis and is a pipeline available
on a web server at: http://ieg2.ou.edu/MENA/main.cgi
It uses random matrix theory to threshold the all-to-all similarity matrix and
offers a number of network analysis options once the network is constructed.
You can find details in
BMC Bioinformatics 2012, 13:113 .
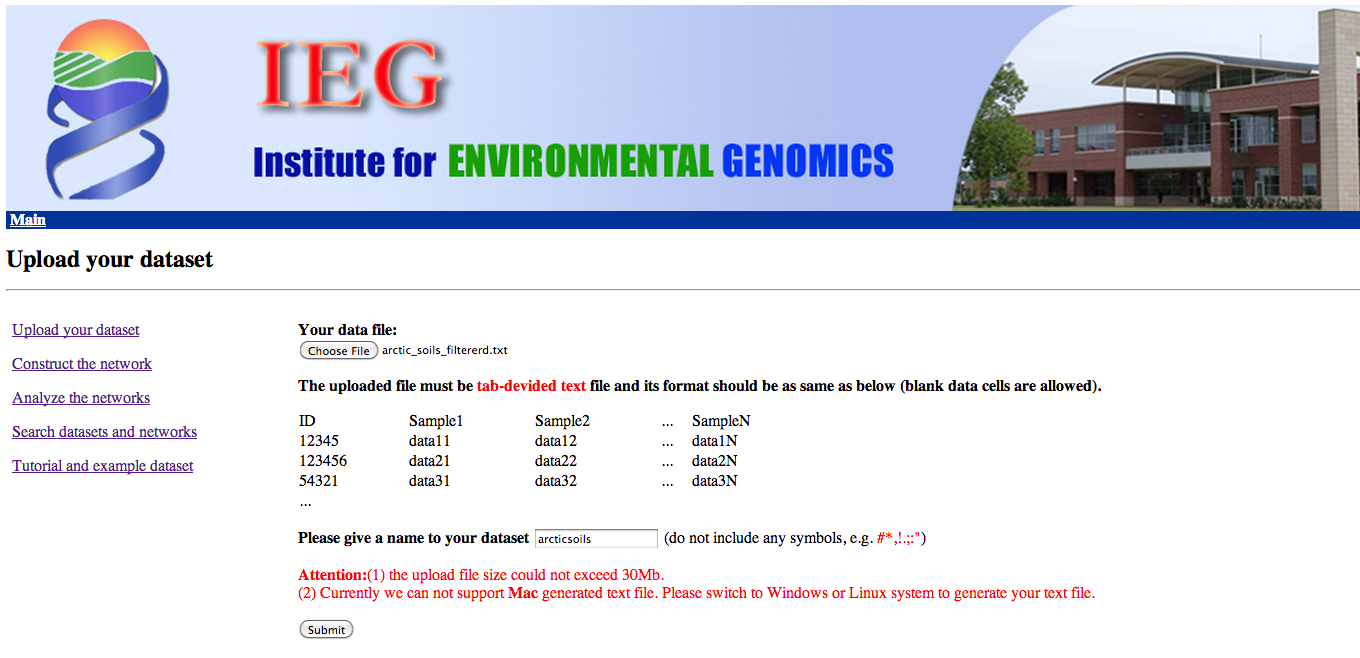
Step 1 - Data upload
Click on "Upload your dataset" -> "Choose file" to upload your data file.
Please choose the input file "arctic_soils_filtered.txt",
where the output from the biom converter has been formatted into a simple OTU count matrix.
Counts of OTUs with less than 20 occurrences were lumped into a single row.
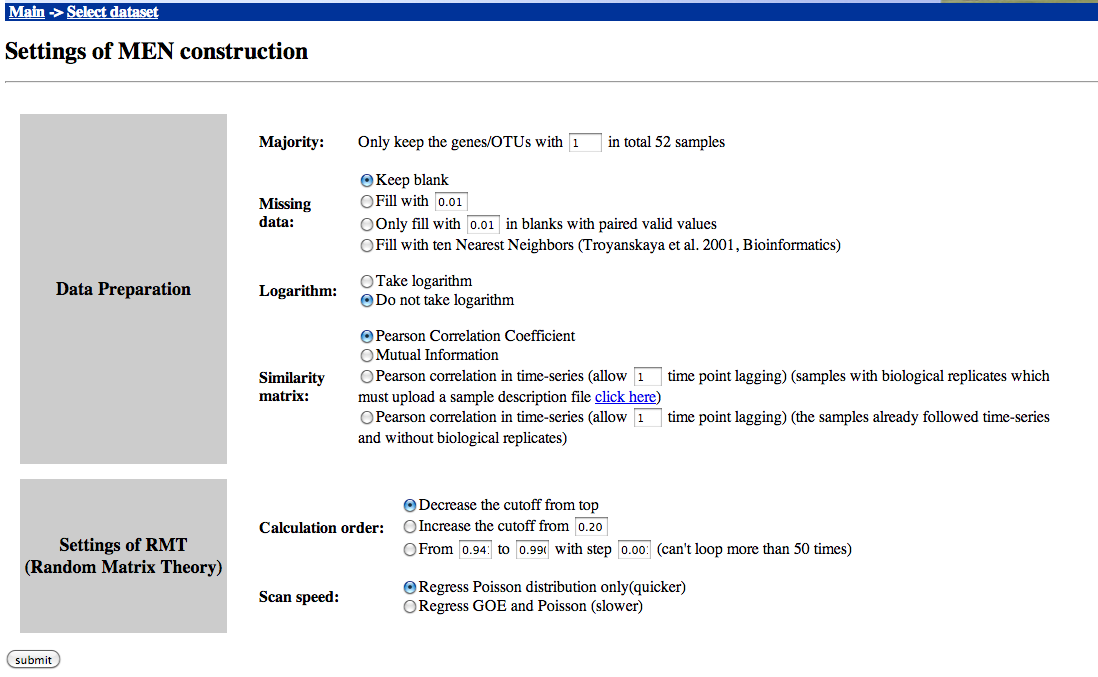
Step 2 - Network construction
You can run MENA's network construction with default values, except for two settings:
Set Majority to 1 (the rare OTUs are already filtered) and choose "Do not take logarithm".
While network construction is running, you can go to the Main window, select "Search
datasets and networks" and check the status of the network construction job. Once it is finished,
click its name in the table and confirm the default threshold proposed by MENA.
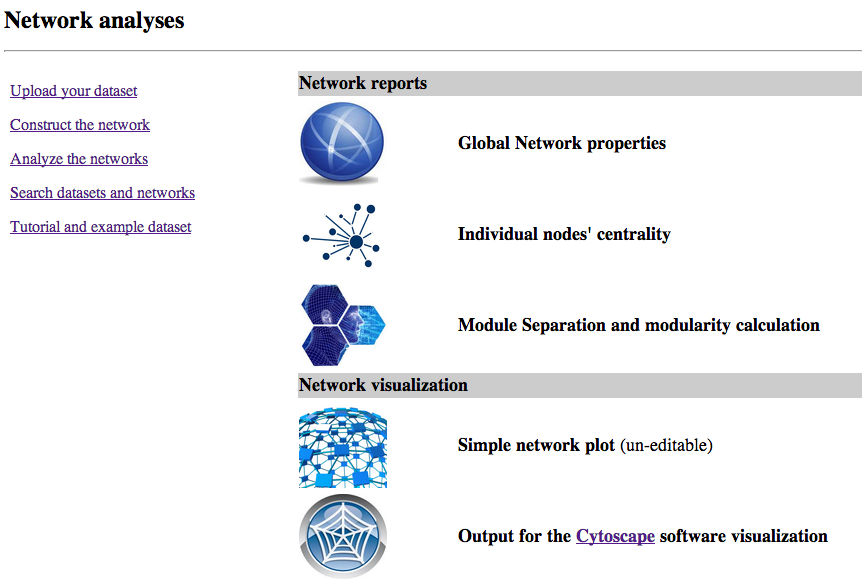
Step 3 - Network analysis
MENA proposes a large number of network analysis options (not all of which are
shown in the screenshot). We will carry out the analyses proposed under "Network reports",
which are required for network download. For this, click each of the symbols in the
"Network reports" section and carry out the analysis with the default values.
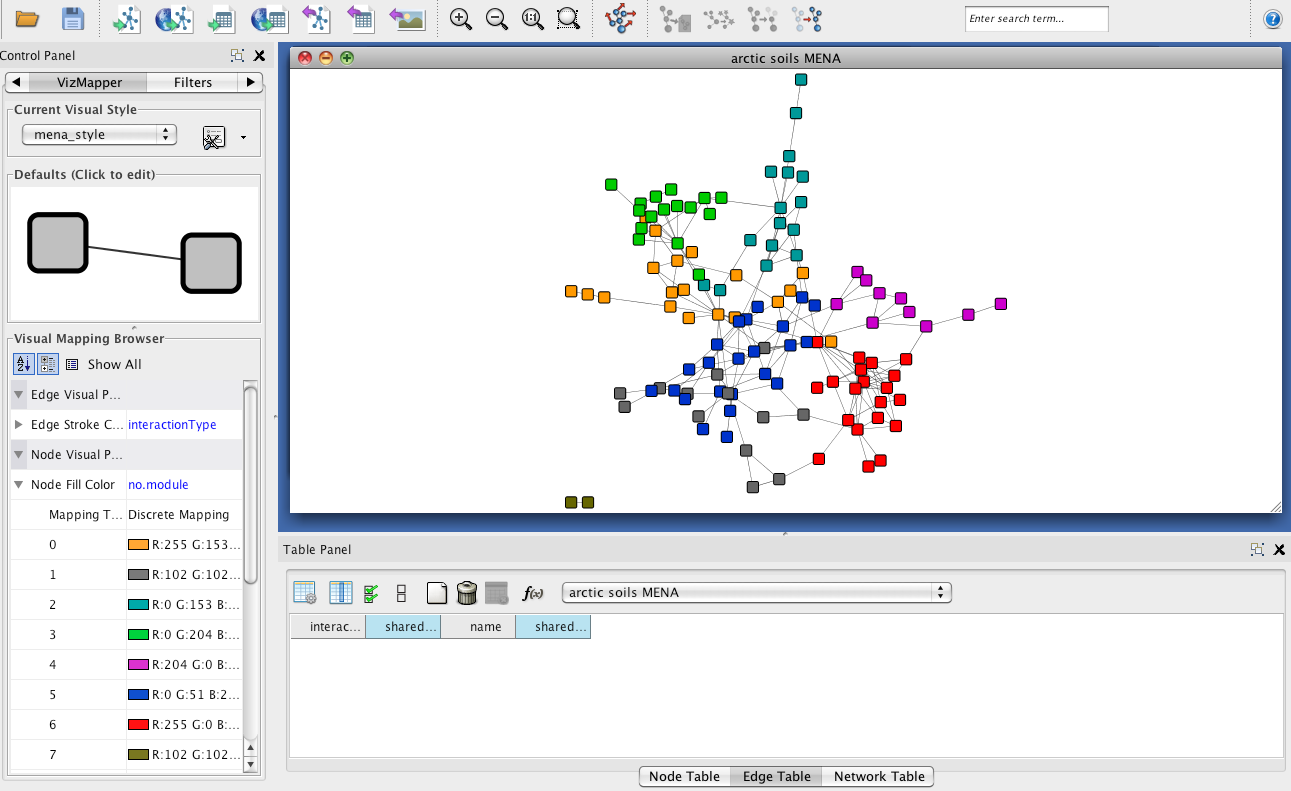
Step 4 - Network visualization
Once network analysis has been carried out, the network can be downloaded
by clicking "Output for the Cytoscape software" in the network analysis menu.
Download the sif file as well as the node attributes and open Cytoscape.
Before loading the MENA network, replace all underscores by dashes in the
sif file as well as in the node attribute file using a simple text editor.
In addition, replace all instances of np by pp in the sif file.
This step will allow comparing the MENA network with the CoNet network later on.
You can then load the sif file via File->Import->Network->File.
Once the network is created, you can load the node attributes using
File->Import->Table->File. You can rename the columns by right-clicking the column name.
Alternatively, you can also click on "Show Text File Import Options" and enable
"Transfer first line as column names".
Make sure to assign the node attributes to the right network collection.
Then, you can choose a network layout (Layout->yFiles Layouts->Organic is recommended).
In the VizMapper, you can first create a MENA-specific style by clicking on the options
next to the current visual style and selecting "Copy visual style". You can name the new
style e.g. "mena_style". You can then select different node colors for the different modules
using the "Node Fill Color" property with the "no.module" node attribute and the Discrete Mapper.